*This is a special feature from the third, Darwin themed print issue of Anthropology Now.*
spitting image, spit’n’ image. Informal. exact likeness; … bef. 950; (v.) ME spitten, OE spittan; c. G (dial.) spitzen to spit; akin to OE spætan to spit, spætl spittle …. (Dictionary. com 2009).
Last year, the California-based project 23andMe—a project that offers to estimate a person’s predisposition for a number of traits and diseases on the basis of a saliva test—held a “spit party” during New York fashion week; volunteers would spit into a test tube to provide their DNA for sequencing and analysis. The photo shows the vibrant scene, a young couple opening their kits and donating saliva, to explore what their genomic constitution might tell them about their identity and the kind of life they might lead. Apparently, they were publicly celebrating both their self and their genome, staging their persona and their bodily essence for the media, in the process of lobbying for personal genomics and the company responsible for 23andMe, an affiliate of Google. An article in The New York Times announced the launching event by saying that 23andMe “wants people to think of their genomes as a basis for social networking,” adding that “the company … hopes to make spitting into a test tube as stylish as ordering a ginger martini” (Salkin 2008). In November 2008, Time Magazine declared the retail DNA test of 23andMe the best innovation of the year. The year before, Apple’s iPhone was the winner. Several other companies have either started or scheduled one form or another of retail genomics. This is consuming genomics, a rapidly growing business receiving both substantial financial support and intense public attention. Clearly, something new is in the air.
The notion of spitting and related concepts has proved to be a powerful metaphor. Exploring its social history is like fol¬lowing the trajectories of ancient DNA. The English verb to “spit”—to “spew” or to “expel saliva”—is of early medieval origin. The noun “spit,” in the meaning “the very likeness,” is more recent, attested from 1602, while “spitting image” is a twentieth-century thing, apparently from as early as 1901. It may be interesting to note that there has been some debate on the etymology of the phrase. Some have suggested it is derived from “splitting image,” based on the two identical parts of a split plank of wood. Such an account would resonate perfectly with the modern concept of the double he¬lix and the splitting of DNA, underlining the relevance of the idea of the “spitting image” for both modern gene talk and the genealogical tree. The discovery of the structure of DNA material has been heralded as the key to the understanding of the continuity and change of life forms, as the missing conceptual link of evolutionary theory finally solving the mystery of the “tree of life.” Also, “splitting” might highlight the Western notion of the duality of the individual as a natural body and a social person, a notion often challenged nowadays by the monistic concept of the biosocial (Rabinow 1996). Given such reasoning, the 23andMe “spit party” might just as well have been called a “split party.” It seems, however, that the reference to spitting was based on “spit,” not “split,” an allusion to someone who is so similar to another as to appear to have been spat out of his or her mouth (Martin 2009).
The spit party and the notion of spitting image invite interesting anthropological questions: What are the overall spin-offs from personal genomics, especially with respect to the understanding of self, person¬hood, relationships, and ancestry? Despite sustained criticisms of the gene talk current in the West and the determinisms it implies, personal genomics along the lines of 23andMe seems to have a substantial public appeal. At the same time, the services offered by genomics companies give rise to new kinds of relations and networks based on genetic signatures presumed to be en¬coded in DNA. Like many others, I decided to indulge in a kind of spitting, mixing ethnographic observation, theoretical reflection, and narcissistic pleasure. One of the key companies in the development of personal genomics, deCODE genetics, hap¬pens to be located on the outskirts of my campus in Reykjavik, Iceland—within spit¬ting distance, if you like.
deCODEme: “Dig into Your DNA!”
A few days before the launching of 23andMe, deCODE genetics announced a similar service—deCODEme. The project now offers both a “complete” scan ($985) and two more narrow scans focusing on specific conditions, cancer ($225) and cardiovascular problems ($195). I signed up for the complete scan, eager to find out how anthropological understanding of humans and their differences was being used and developed in the project, to explore the assumptions about cultures and bodies on which analyses would be based, to see what the scan might tell me about myself and my roots, and to follow the development of the virtual community of people who subscribe to services of this kind. For some months I resisted the narcissism of personal genomics. Both of my parents had struggled with cancer and I wasn’t terribly keen on the kind of fortune telling offered by personal genomics. I guess news of the New York spit party helped to change my mind. Somehow collective spitting and the bonding involved appealed to the anthropologist, curious about the implications of the new genetics for modern life. In my case, how¬ever, there was no formal party. Extracting the cheek swabs, signing the relevant forms, and mailing the lot to the lab was a solitary event.
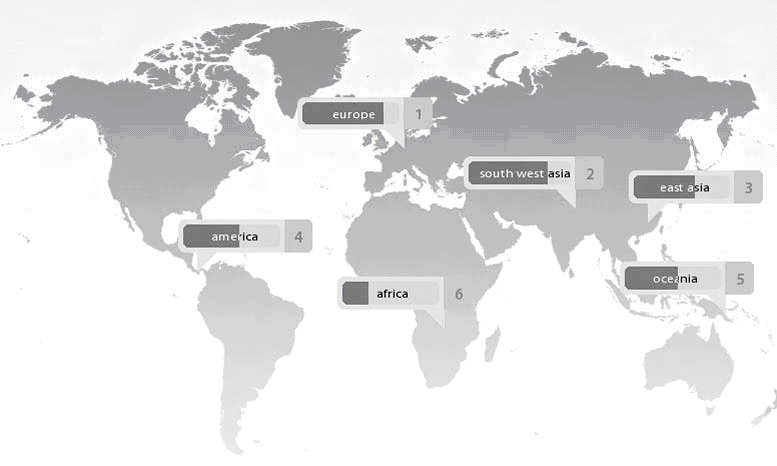
Two weeks later, I received an email from the company. The results were now available and I would be able to access them through the password provided. Once I logged on, I was urged to “have fun browsing [my] … genome,” “dig into [my] … DNA,” explore my ancestry and my “genetic risks,” play with fancy maps and other visuals, search for specific genetic variants (SNPs or “snips”), and download my genotypes for 1.2 million SNPs (a 33Mb datafile). The comparison of my genetic code with that of populations covered in the “Genetic Atlas,” I was told, was based on several hundred thousand genetic variants and more than 1,000 reference individuals from 50 different populations worldwide (see the illustration). My genome, not surprisingly, turned out to have most in common with “European” reference groups (a genetic similarity of 83.99%), in particular those of Iceland, the Orkneys, France, and Russia.
More astonishingly, another feature of deCODEme, “ancestral origins,” indicated that judging from chromosomes 1 to 22 my ancestry was no less than 7% East Asian, 16% according to the X chromosome, considerably higher than for most Icelanders. I found this an interesting and puzzling revelation. To speak of “genealogical dis-ease” (Rapp, Heath, and Taussig 2001)—to use a term developed by anthropologists studying what people make of genetic information about their roots and ancestry—would, however, be an overstatement.
According to the analysis of my maternal DNA, I belong to “mitogroup R*.” This is a category shared by 4.8% of deCODEme users, all of whom can trace their mitochondrial DNA to a woman thought to have lived about 60 thousand years ago, probably somewhere in the Near East. Analysis of my paternal DNA, on the other hand, shows that I belong to “Y-group R1a,” a category shared with 10.3% of deCODEme users tracing their Y chromosomes back to one man who is thought to have lived about 10 to 15 thousand years ago, probably in Western Asia. A further feature allows users to explore their “map of kinship,” a visual representation of genetic space on the basis of so-called principal component analysis (PCA, for short). This method compares the genetic code of many individuals to uncover genetic patterns or dimensions involving many different SNPs. On the basis of this evidence, I seem to occupy a somewhat marginal position, neither firmly within the European reference group nor any of the others, probably reflecting the puzzling observation about my partial East Asian ancestry.
The other main service offered by de-CODEme is that of analyzing the genome with respect to specific traits and health risks. The current list of risks analyzed by deCODEme is a mixed bag of forty diseases and traits, including alcohol flush reaction, Alzheimer’s disease, heart attack, lactose in¬tolerance, male pattern baldness, multiple sclerosis, prostate cancer, and psoriasis. My results for the diseases and traits covered are based on calculations comparing my genetic sequence to sequences of participants in studies published in the scholarly literature. To access results for some diseases I was invited to read about the genetic and medical details and to sign a statement about informed consent, by clicking on “Accept.”
I need not bore the reader with the personal details. Suffice it to say that some of the information provided sounds trivial (no alcohol flush reaction), some of it resonates with what I thought I already knew (I am less likely than the general population “to become nicotine dependent [15% or less]”), some results are encouraging (I have low lifetime risks for some diseases, much less than for males of European ancestry in general), and some details may encourage the hypochondriac in me to request further medical information (my risks for some dis¬eases are significantly higher than those of my genetically significant others).
When presented with these results, I was offered details on the mathematics of risk analysis. Also, I was invited to zoom in on my genomic landscape, focusing on a part of a chromosome and the location of specific mutations reportedly responsible for potential traits or diseases. The website drew the parallels of the two universes of in¬side and outside: “In the same manner as Google Earth allows you to explore the world map, the deCODEme Genome Browser enables you to visualize the genome.” The tour was far more fun than the mathematics. Again, there were some surprises and some food for thought.
Emergent Communities and Technologies of the Self
It seems reasonable to argue, as Hacking observes (2009), that personal genomics represent one example of what Michel Foucault referred to as “technologies of the self.” For Foucault, technologies of the self “permit individuals to effect by their own means or with the help of others a certain number of operations on their own bodies and souls, thoughts, conduct, and a way of being, so as to transform themselves in order to attain a certain state of happiness, purity, wisdom, perfection, or immortality” (1988:18). One of the pioneers of the genetics of ancestry is Bryan Sykes of Oxford University. Significantly, his book on ancestry (2001) opens with the question “Where do I come from?” and closes with a chapter on “A Sense of Self.”
For many people, then, hereditary material provides an important avenue into identity and personhood. Knowing our genetic constitution and where we come from, we apparently also know who we are. As Pinker observes (2009): “Affordable genotyping may offer new kinds of answers to the question ‘Who am I?’—our ruminations about our ancestry, our vulnerabilities, our character and our choices in life.” This is a theme underlined by Anne Wojcicki, the cofounder of 23andMe: the 600,000 genetic markers interpreted by 23andMe, she argues, are “the digital manifestation of you” (see Hamilton 2008).
One may object to the rhetoric of self-discovery evident in the marketing of personal genomics by saying that personhood is not a matter of genetics, whatever people say at modern spit parties. Indeed, many ethnographies would testify to other ways of establishing personhood. For many Canadian Inuit, for instance, personhood is largely framed in the context of name talk, not gene talk (Pálsson 2008); the essence of the person, it is assumed, is constructed through a highly communal project heavily dependent on personal naming. During the life course, a person acquires a series of recycled names from friends and relatives that collectively establish the identity, personality, and fate of the individual concerned. An Inuit spit party, as a result, is likely to have a radically different meaning from that of many New York spit parties. Perhaps one should keep in mind that gene talk has only been around for half a century or so while name talk has probably followed culture since the beginning of humanity. New York celebrities, however, much like Inuit and everybody else, construct their personhood and identities in the course of everyday life, possibly through “Facebook” kinds of networking based on genome sequencing. The virtual becomes the real thing.
Personal genomics not only establishes, it is assumed, who we are, but it also generates new networks and communities. Indeed, a thriving imagined community of the users of personal genomics projects has been developing on the Internet. The Genome Browser of deCODEme allows users to compare their complete data with friends and family. While my reference group of friends and family includes both hypochondriacs and anthropologists, so far they have seen few good reasons to participate and, as a result, there isn’t much to compare. The Web site, however, allowed me to examine my genetic sharing with three “famous” people: Kári Stefánsson (the president, CEO, and cofounder of deCODE genetics), Craig Venter (founder of the Institute for Genomic Research), and James D. Watson (codiscoverer of the structure of DNA). Here, sharing is indicated visually by the coloring of the relevant bits of the chromosomes. Not surprisingly, I had more in common with my fellow Icelander than with Venter and Watson. So, after all, I did have a kind of spit party, online in absentia.
No doubt personal genomics is becoming both a family affair and a global concern. A number of websites testify to a lively discourse on the issues involved, including thinkgene.com, dna-forums.org, Eye on DNA, Dienekes’ Anthropology Blog, and Urban Semiotics. Users can draw their own conclusions from the analyses provided and engage in dialogues with genomic experts, sometimes becoming experts themselves in the process. Several users have tried two or more services to explore the extent to which their results on health risks and ancestry might agree or disagree. Sometimes people check if they are being cheated. One blogger claimed to know of “at least one case … where a customer deliberately submitted a dog’s DNA just to ‘test’ the company. He was willing to pay for his little experiment, and yes, the company figured out exactly what had happened!”
Many people have little interest in exploring their health risks in public. Judging from the websites, there seems to be more interest in discussing ancestry. Sykes’s service (Oxford Ancestry), which offers people an opportunity to see which “clan” they belong to, to trace their ancestry to one of the seven daughters of Eve, has generated extensive discussion. Here is a reference from one of the blog sites:
I received my DNA results earlier this year and was surprised to find myself in clan Ulrike. I have traced six generations of ma¬ternal ancestors in the Beds/Northants bor¬ders region. The Viking invaders did travel into this area…. I have always been at¬tracted to northern wilderness and have visited Alaska, Greenland/Iceland and Siberia. Is this my DNA speaking?!
Some bloggers take a playful attitude to genome testing. One woman had her husband “tested” for fun:
I admit it. I have no self-discipline when it comes to genetic genealogy. When de-CODEme launched, I had to be one of the first in line to get tested. So I ordered … and received results … —my husband’s results, that is. I thought this might be a little more interesting since he sports a Y chromosome.
Clearly, there is a rapidly growing interest in personal genomics, for the purpose of celebrating our past and for managing our lives and our future.
Analyses of ancestry are likely to remain more or less intact, despite some anthropological doubts about important issues (Bolnick et al. 2007, Marks 2008), including the identification and sampling of populations and the shape of the family tree, partly because there is not so much at stake and, in any case, it is play. Studies of the genomics of diseases, in contrast, are riddled with contests, doubts, and conflict. Most common diseases are only minimally explained by genetic factors and in each case a great number of genes are likely to be involved. Also, the exact constellation of genes seems important, which further complicates analyses. Last, but not least, there is growing evidence for the importance of “epigenetic” factors, way beyond the simple concept of DNA sequence.
Given the evidence, and the growing public awareness of it (see, for instance, Hall 2009), why would people bother to measure their health risks with personal genomics services? While the hype may have faded, there seems to be a continued market for the kinds of services provided. The narcissistic pleasures of late modernity are reaching levels that Foucault could not possibly anticipate, and personal genomics is just one example of the fascination with the body. Also, the power of computing machinery continues to expand and cheap complete sequencing is within reach. Moreover, there are immense financial stakes and concerns on the global level, for biotechnical and pharmaceutical companies. As a result, one may expect personal genomics projects to expand. Although deCODE genetics has been in dire financial shape for some time, burdened with the excessive costs of its scientific work and its laboratory, and its future remains uncertain, other companies specializing in personal genomics seem to thrive. New services continue to be added to the menu.
Laboring Consumers
The companies involved in personal genomics emphasize consumers’ relative autonomy and independence from the medical establishment. Indeed, personal genomics of the kind discussed here may involve an element of empowerment. The virtual community of genetic citizens actively debates and negotiates roots, identities, and health risks fusing the expertise of professional and “lay” geneticists for the purpose of scrutinizing genomes. In a sense, then, this is science from below (Harding 2008). The forums involved are reasonably democratic social networks based on identification with genomic characteristics.
While giving people an opportunity to become active explorers and governors of their genomes is a good idea, the arguments about individual freedom, informed choices, and the unregulated genomic marketplace emphasized by genomic companies should be taken with a grain of salt. For one thing, they disguise the fact, as Prainsack et al. argue (2008: 34), that personal genomics is pushing the individualization of responsibilities a bit too far. Public authorities, they suggest, should “make it a priority to fund empirical research exploring what individuals expect from personal genomics, and in what way genetic susceptibility information is likely to affect practices and lifestyle choices.” Here, anthropology can play an important role (Nelson 2008, Santos et al. 2009).
Another qualification concerns the labor that users of genomics services perform for personal-genomics services. I suggest that genomic services engage the bodies and labor power of their consumers in what may be called biosocial relations of production (Pálsson 2009). The spokespersons for 23andMe, unlike most of the other projects, including deCODEme, have been quite open about the issue of alternative uses of their data. Wojcicki suggests signing up for 23andMe is “a great way for individuals to be involved in the research world….
You will have a profile, and something almost like a ribbon marking participation in these different research papers. It will be like, ‘How many Nature articles have you been part of?’” (pimm.wordpress.com 2007). The people contributing spits and cheek swabs to personal genomics services, then, take part in a labor process that ultimately may result in other projects, including large-scale biobanking. Whatever their current ambitions, personal genomics projects are likely to connect with larger bio¬medical projects in the future. Spitting and snipping, after all, is work, potentially contributing to the global networks and hierarchies involved in the manufacture of biovalue.
Recombinant Metaphors
The image and the report in the New York Times regarding the launching of 23andMe draw attention to the role of metaphors. Reporters quickly drew upon a series of related metaphors; the event was described as a “spit party,” the message of 23andMe and personal genomics in general, it was argued, was “when in doubt, spit it out,” personal medicine was said to be “within spitting distance,” and so on. The people of 23andMe now have a blog site called “The Spittoon,” drawing its name from an object also called “spitter,” a receptacle for spitting into: “Using nothing more than a bit of saliva (Get it? The Spittoon!), the genotyping process we use analyzes more than 580,000 locations in a person’s genome” (The Spittoon 2009). It is tempting to assume that the spit is becoming one of the key metaphors we live by, informing our speech and our thoughts. Metaphors, however, just like DNA, frequently undergo mutations, re¬combining available material from everyday language and experience. The notion of the “spitting image” as we have seen, is a case in point.
While 23andMe is probably the only personal genomics project that uses “spittoon” samples and the others seem generally to draw upon buccal swabs, the “spit party” seems to nicely capture various aspects of personal genomics. It captures the gene talk on which it is based, the mechanisms of inheritance, the matching or mismatching disclosed through the sequencing of DNA material, and the establishing of distance and ancestry, both genetic and social. When spitting out one’s saliva, one is presumed to provide a spitting image of oneself, encoded in DNA. The transparent metaphor has, finally, been elevated above the debates of etymologists. The emphasis, on the other hand, is no longer on spitting at someone (usually a gesture of contempt) but on the conviviality of spitting with a fellow human being, for the purpose of celebrating biosocial bonds, for founding social networks based on bodily signatures—with a ginger martini!
References
Bolnick, Deborah A., et al. 2007. “The Science and Business of Genetic Ancestry Testing.” Science 318 (19 October): 399–400.
Dictionary.com. 2009. http://dictionary.reference. com/browse/. Accessed 29 May.
Foucault, Michel. 1988. “Technologies of the Self.” In Luther H. Martin, Huck Gutman, and Patrick H. Hutton, eds. Technologies of the Self. 16–50. Amherst, MA: The University of Massachusetts Press.
Hacking, Ian. 2009. “What Will Commercial Genome-Reading—from Cheap 23andMe to Costly but Complete Knome—Do to Middle-Class Conceptions of Personal Identity? On the Human Forum: Current Controversies in the Study of Humans, Animals, and Machines. http:// onthehuman.org/humannature/?p=176&cpage= 1#comment-295. 30 March.
Hall, Stephen S. 2009. “Beyond the Book of Life.” Newsweek, July 6/July 13: 38–41.
Hamilton, Anita. 2008. “The Retail DNA Test.” Time Magazine. November 3. http://www.time. com/time/specials/packages/article/0,28804 ,1852747_1854493,00.html.
Harding, Sandra. 2008. Sciences from Below: Feminisms, Postcolonialities, and Modernities. Durham, NC: Duke University Press.
Marks, Jonathan. 2008. “Recreational Ancestry-Caveat Emptor? Relatedness Is More Complex Than Commercial Gene-Based Family Trees Would Suggest.” Genetic Engineering & Biotechnology News 21, no. 11.
Martin, Gary. 2009. “Spitting Image.” The Phrase Finder. www.phrases.org.uk/meanings/spitting -image.html. Accessed 11 May.
Nelson, Alondra. 2008. “Bio Science: Genetic Genealogy Testing and the Pursuit of African Ancestry.” Social Studies of Science 38, no. 5: 759–783.
Pálsson, Gísli. 2008. “Genomic Anthropology: Coming in from the Cold?” Current Anthropology 49, no. 4:545–568.
———. 2009. “Biosocial Relations of Production.” Comparative Studies in Society and His¬tory 51, no. 2:288–313.
pimm.wordpress.com. 2007. “23andMe’s Mission: Connecting All People on the DNA Level or Social Networking XY.0.” November 19.
Pinker, Stephen. 2009. “My Genome, My Self.” The New York Times, January 11.
Prainsack, Barbara, et al. 2008. “Misdirected Precaution.” Nature 456 (6): 34–35.
Rabinow, Paul. 1996. Essays on the Anthropology of Reason. Princeton: Princeton University Press.
Rapp, Rayna, Deborah Heath, and Karen-Sue Taussig. 2001. “Genealogical Dis-ease: Where Hereditary Abnormality, Biomedical Explanation, and Family Responsibility Meet.” In Sarah Franklin and Susan McKinnon, eds. Relative Values: Reconfiguring Kinship Studies. Durham, NC: Duke University Press, 384–412.
Santos, Ricardo Ventura, et al. 2009. “Color, Race and Genomic Ancestry in Brazil: Dialogues between Anthropology and Genetics.” Current Anthropology. In press.
Salkin, Allen. 2008. “When in Doubt, Spit It Out.” The New York Times, 12 September.
The Spittoon. 2009. http://spittoon.23andme. com/. Accessed 29 May.
Sykes, Bryan. 2001. The Seven Daughters of Eve. London: Bantam Press.
Gísli Pálsson is professor, Department of Anthro¬pology, Gimli, University of Iceland, 101 Reykjavik, Iceland. His most recent book is Anthropology and the New Genetics (2007). Currently, Pálsson’s research focuses on the social implications of biotechnology, genetic history, and environmental change.